Programming in R
31 minute read
Overview
One of the main attractions of using the R (http://cran.at.r-project.org) environment is the ease with which users can write their own programs and custom functions. The R programming syntax is extremely easy to learn, even for users with no previous programming experience. Once the basic R programming control structures are understood, users can use the R language as a powerful environment to perform complex custom analyses of almost any type of data (Gentleman 2008).
Why Programming in R?
- Powerful statistical environment and programming language
- Facilitates reproducible research
- Efficient data structures make programming very easy
- Ease of implementing custom functions
- Powerful graphics
- Access to fast growing number of analysis packages
- One of the most widely used languages in bioinformatics
- Is standard for data mining and biostatistical analysis
- Technical advantages: free, open-source, available for all OSs
R Basics
The previous Rbasics tutorial provides a general introduction to the usage of the R environment and its basic command syntax. More details can be found in the R & BioConductor manual here.
Code Editors for R
Several excellent code editors are available that provide functionalities like R syntax highlighting, auto code indenting and utilities to send code/functions to the R console.
- RStudio: GUI-based IDE for R
- Vim-R-Tmux: R working environment based on vim and tmux
- Emacs (ESS add-on package)
- gedit and Rgedit
- RKWard
- Eclipse
- Tinn-R
- Notepad++ (NppToR)
Finding Help
Reference list on R programming (selection)
- Advanced R, by Hadley Wickham
- R Programming for Bioinformatics, by Robert Gentleman
- S Programming, by W. N. Venables and B. D. Ripley
- Programming with Data, by John M. Chambers
- R Help & R Coding Conventions, Henrik Bengtsson, Lund University
- Programming in R (Vincent Zoonekynd)
- Peter’s R Programming Pages, University of Warwick
- Rtips, Paul Johnsson, University of Kansas
- R for Programmers, Norm Matloff, UC Davis
- High-Performance R, Dirk Eddelbuettel tutorial presented at useR-2008
- C/C++ level programming for R, Gopi Goswami
R Scripts
R programming involves the writing of R scripts, which are text files containing R code. The file name of an R script carries the extension .R (e.g. myscript.R). Typically, R scripts are composed of code sections, as well as comments (starting with #) explaining what the code does.
The structure and styling requirements of R scripts are not very strict. However, for maintainability and readability, it is highly recommended to follow widely adopted styling recommendations. For details on how to best and consistently format R scripts, this style guide is a good start. In addition, the formatR package is very helpful.
In addition to executing the code in R scripts interactively, e.g. line by line, one can execute R scripts as a whole with execution commands from the R console or the command-line using source or Rscript, respectively. The details on how to execute R scripts are explained in this section.
R Markdown (Rmd) and Quarto (qmd) scripts are an extension of R scripts that include in addition to R code, results and commentary text. Both can be rendered to a variety formats including HTML and PDF. They are explained in a separate tutorial here. R packages are often built for organizing more complex R code and providing usage instructions in the form of help files and vignettes (manuals). How to build R packages is explained in the R Package tutorial of this website.
The code in R scripts (or Rmd/qmd scripts) should be organized as much as possible in R functions that are called (used) somewhere in the script. Alternatively, functions can be organized in a secondary R script from where they are imported with the source command. This makes the code more concise, readable and reusable.
Control Structures
Important Operators
Comparison operators
==(equal)!=(not equal)>(greater than)>=(greater than or equal)<(less than)<=(less than or equal)
Logical operators
&(and)&&(and)|(or)||(or)!(not)
Note: & and && indicate logical AND, while | and || indicate logical OR. The shorter form performs element-wise comparisons of same-length vectors.
The longer form evaluates left to right examining only the first element of each vector (can be of different lengths). Evaluation proceeds only until the result
is determined. The longer form is preferred for programming control-flow, e.g. via if clauses.
Conditional Executions: if Statements
An if statement operates on length-one logical vectors.
Syntax
if (TRUE) {
statements_1
} else {
statements_2
}
In the else component, avoid inserting newlines between } else.
Example
if (1==0) {
print(1)
} else {
print(2)
}
## [1] 2
Example 2
if (1==0) {
print(1)
} else if (1==2) {
print(2)
} else {
print(3)
}
## [1] 3
Conditional Executions: ifelse Statements
The ifelse statement operates on vectors.
Syntax
ifelse(test, true_value, false_value)
Example
x <- 1:10
ifelse(x<5, sqrt(x), 0)
## [1] 1.000000 1.414214 1.732051 2.000000 0.000000 0.000000 0.000000 0.000000 0.000000 0.000000
Loops
for loop
for loops iterate over elements of a looping vector.
Syntax
for(variable in sequence) {
statements
}
Example
mydf <- iris
myve <- NULL
for(i in seq(along=mydf[,1])) {
myve <- c(myve, mean(as.numeric(mydf[i,1:3])))
}
myve[1:8]
## [1] 3.333333 3.100000 3.066667 3.066667 3.333333 3.666667 3.133333 3.300000
Note: Inject into objecs is much faster than append approach with c, cbind, etc.
Example
myve <- numeric(length(mydf[,1]))
for(i in seq(along=myve)) {
myve[i] <- mean(as.numeric(mydf[i,1:3]))
}
myve[1:8]
## [1] 3.333333 3.100000 3.066667 3.066667 3.333333 3.666667 3.133333 3.300000
Conditional Stop of Loops
The stop function can be used to break out of a loop (or a function) when a condition becomes TRUE. In addition, an error message will be printed.
Example
x <- 1:10
z <- NULL
for(i in seq(along=x)) {
if (x[i] < 5) {
z <- c(z, x[i]-1)
print(z)
} else {
stop("values need to be < 5")
}
}
while loop
Iterates as long as a condition is true.
Syntax
while(condition) {
statements
}
Example
z <- 0
while(z<5) {
z <- z + 2
print(z)
}
## [1] 2
## [1] 4
## [1] 6
The apply Function Family
apply
Syntax
apply(X, MARGIN, FUN, ARGs)
Arguments
X:array,matrixordata.frameMARGIN:1for rows,2for columnsFUN: one or more functionsARGs: possible arguments for functions
Example
apply(iris[1:8,1:3], 1, mean)
## 1 2 3 4 5 6 7 8
## 3.333333 3.100000 3.066667 3.066667 3.333333 3.666667 3.133333 3.300000
tapply
Applies a function to vector components that are defined by a factor.
Syntax
tapply(vector, factor, FUN)
Example
iris[1:2,]
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
tapply(iris$Sepal.Length, iris$Species, mean)
## setosa versicolor virginica
## 5.006 5.936 6.588
sapply, lapply and vapply
The iterator functions sapply, lapply and vapply apply a function to
vectors or lists. The lapply function always returns a list, while sapply
returns vector or matrix objects when possible. If not then a list is
returned. The vapply function returns a vector or array of type matching the
FUN.VALUE. Compared to sapply, vapply is a safer choice with respect to
controlling specific output types to avoid exception handling problems.
Examples
l <- list(a = 1:10, beta = exp(-3:3), logic = c(TRUE,FALSE,FALSE,TRUE))
lapply(l, mean)
## $a
## [1] 5.5
##
## $beta
## [1] 4.535125
##
## $logic
## [1] 0.5
sapply(l, mean)
## a beta logic
## 5.500000 4.535125 0.500000
vapply(l, mean, FUN.VALUE=numeric(1))
## a beta logic
## 5.500000 4.535125 0.500000
Often used in combination with a function definition:
lapply(names(l), function(x) mean(l[[x]]))
sapply(names(l), function(x) mean(l[[x]]))
vapply(names(l), function(x) mean(l[[x]]), FUN.VALUE=numeric(1))
Programming Exercise on Loops: go here
Improving Speed Performance of Loops
Looping over very large data sets can become slow in R. However, this
limitation can be overcome by eliminating certain operations in loops or
avoiding loops over the data intensive dimension in an object altogether. The
latter can be achieved by performing mainly vector-to-vecor or
matrix-to-matrix computations. These vectorized operations run in R often over
100 times faster than the corresponding for() or apply() loops. In
addition, one can make use of the existing speed-optimized C-level functions in
R, such as rowSums, rowMeans, table, and tabulate. Moreover, one can
design custom functions that avoid expensive R loops by using vector- or
matrix-based approaches. Alternatively, one can write programs that will
perform all time consuming computations on the C-level.
The following code samples illustrate the time-performance differences among the different approaches of running iterative operations in R.
1. for loop with append versus inject approach
The following runs a for loop where the result is appended in each iteration
with the c() function. The corresponding cbind and rbind for two dimensional
data objects would have a similar performance impact as c().
myMA <- matrix(rnorm(1000000), 100000, 10, dimnames=list(1:100000, paste("C", 1:10, sep="")))
results <- NULL
system.time(for(i in seq(along=myMA[,1])) results <- c(results, mean(myMA[i,])))
user system elapsed
39.156 6.369 45.559
Now the for loop is run with an inject approach for storing the results in each iteration.
results <- numeric(length(myMA[,1]))
system.time(for(i in seq(along=myMA[,1])) results[i] <- mean(myMA[i,]))
user system elapsed
1.550 0.005 1.556
As one can see from the output of system.time, the inject approach is 20-50 times faster.
2. apply loop versus rowMeans
The following performs a row-wise mean calculation on a large matrix first with an apply
loop and then with the rowMeans function.
system.time(myMAmean <- apply(myMA, 1, mean))
user system elapsed
1.452 0.005 1.456
system.time(myMAmean <- rowMeans(myMA))
user system elapsed
0.005 0.001 0.006
Based on the results from system.time, the rowMeans approach is over 200 times faster
than the apply loop.
3. apply loop versus vectorized approach
In this example row-wise standard deviations are computed with an apply loop and then
in a vectorized manner.
system.time(myMAsd <- apply(myMA, 1, sd))
user system elapsed
3.707 0.014 3.721
myMAsd[1:4]
1 2 3 4
0.8505795 1.3419460 1.3768646 1.3005428
system.time(myMAsd <- sqrt((rowSums((myMA-rowMeans(myMA))^2)) / (length(myMA[1,])-1)))
user system elapsed
0.020 0.009 0.028
myMAsd[1:4]
1 2 3 4
0.8505795 1.3419460 1.3768646 1.3005428
The vector-based approach in the last step is over 200 times faster than the apply loop.
4. Example of fast querying routine applied to a large matrix
(a) Create a sample matrix
The following lfcPvalMA function creates a test matrix containing randomly generated log2 fold changes (LFCs)
and p-values (here: pval or FDRs) for variable numbers of samples or test results. In biology this dataset mimics the
results of an analysis of differentially expressed genes (DEGs) from several contrasts arranged in a
single matrix (or data.frame).
lfcPvalMA <- function(Nrow=200, Ncol=4, stats_labels=c("lfc", "pval")) {
set.seed(1410)
assign(stats_labels[1], runif(n = Nrow * Ncol, min = -4, max = 4))
assign(stats_labels[2], runif(n = Nrow * Ncol, min = 0, max = 1))
lfc_ma <- matrix(lfc, Nrow, Ncol, dimnames=list(paste("g", 1:Nrow, sep=""), paste("t", 1:Ncol, "_", stats_labels[1], sep="")))
pval_ma <- matrix(pval, Nrow, Ncol, dimnames=list(paste("g", 1:Nrow, sep=""), paste("t", 1:Ncol, "_", stats_labels[2], sep="")))
statsMA <- cbind(lfc_ma, pval_ma)
return(statsMA[, order(colnames(statsMA))])
}
degMA <- lfcPvalMA(Nrow=200, Ncol=4, stats_labels=c("lfc", "pval"))
dim(degMA)
## [1] 200 8
degMA[1:4,] # Prints first 4 rows of DEG matrix generated as a test data set
## t1_lfc t1_pval t2_lfc t2_pval t3_lfc t3_pval t4_lfc t4_pval
## g1 -1.8476368 0.39486484 1.879310 0.7785999 0.1769551 0.9904342 0.1747932 0.9536679
## g2 0.2542926 0.04188993 -1.629778 0.6379570 -1.9280792 0.6106041 -2.3599518 0.1950022
## g3 3.4703657 0.73881357 2.047794 0.3129176 -3.8891714 0.1508787 3.7811606 0.2560303
## g4 -2.8548158 0.99201512 -2.710385 0.1772805 1.0920515 0.2826038 3.9313225 0.5519854
(b) Organize results in list
To filter the results efficiently, it is usually best to store the two different
stats (here lfc and pval) in separate matrices (here two) where each has the
same dimensions and row/column ordering. Note, in this case a list
is used to store the two matrices.
degList <- list(lfc=degMA[ , grepl("lfc", colnames(degMA))], pval=degMA[ , grepl("pval", colnames(degMA))])
names(degList)
## [1] "lfc" "pval"
sapply(degList, dim)
## lfc pval
## [1,] 200 200
## [2,] 4 4
(c) Combinatorial filter
With the above generated data structure of two complementary matrices it is
easy to apply combinatorial filtering routines that are both flexible and
time-efficient (fast). The following example queries for fold changes of at
least 2 (here lfc >= 1 | lfc <= -1) plus p-values of 0.5 or lower. Note, all
intermediate and final results are stored in logical matrices. In addition to
boolean comparisons, one can apply basic mathematical operations, such as
calculating the sum of each cell across many matrices. This returns a numeric matix of
integers representing the counts of TRUE values in each position of the
considered logical matrices. Subsequently, one can perform summary and
filtering routines on these count-based matrices which is convenient when
working with large numbers of matrices. All these matrix-to-matrix comparisons
are very fast to compute and require zero looping instructions by the user.
queryResult <- (degList$lfc <= 1 | degList$lfc <= -1) & degList$pval <= 0.5
colnames(queryResult) <- gsub("_.*", "", colnames(queryResult)) # Adjust column names
queryResult[1:4,]
## t1 t2 t3 t4
## g1 TRUE FALSE FALSE FALSE
## g2 TRUE FALSE FALSE TRUE
## g3 FALSE FALSE TRUE FALSE
## g4 FALSE TRUE FALSE FALSE
(d) Extract query results
- Retrieve row labels (genes) that match the query from the previous step in each column, and
store them in a
list.
matchingIDlist <- sapply(colnames(queryResult), function(x) names(queryResult[queryResult[ , x] , x]), simplify=FALSE)
matchingIDlist
## $t1
## [1] "g1" "g2" "g5" "g6" "g11" "g16" "g18" "g19" "g21" "g23" "g24" "g31" "g36"
## [14] "g37" "g41" "g46" "g60" "g61" "g63" "g70" "g71" "g72" "g75" "g81" "g83" "g84"
## [27] "g88" "g91" "g97" "g98" "g100" "g102" "g103" "g104" "g110" "g111" "g112" "g113" "g114"
## [40] "g120" "g121" "g123" "g124" "g126" "g130" "g134" "g135" "g139" "g140" "g145" "g147" "g153"
## [53] "g157" "g158" "g159" "g160" "g162" "g170" "g171" "g172" "g173" "g175" "g178" "g183" "g184"
## [66] "g187" "g190" "g192" "g196" "g199"
##
## $t2
## [1] "g4" "g5" "g7" "g9" "g10" "g12" "g16" "g23" "g34" "g35" "g39" "g41" "g44"
## [14] "g46" "g47" "g48" "g49" "g50" "g51" "g52" "g56" "g66" "g75" "g80" "g81" "g85"
## [27] "g88" "g89" "g90" "g94" "g99" "g102" "g112" "g115" "g116" "g118" "g119" "g120" "g129"
## [40] "g144" "g145" "g148" "g152" "g155" "g156" "g160" "g164" "g165" "g167" "g168" "g170" "g172"
## [53] "g178" "g186" "g187" "g194" "g197"
##
## $t3
## [1] "g3" "g6" "g7" "g9" "g12" "g15" "g23" "g25" "g26" "g27" "g38" "g43" "g52"
## [14] "g53" "g54" "g58" "g66" "g69" "g72" "g76" "g77" "g80" "g84" "g85" "g86" "g88"
## [27] "g89" "g90" "g91" "g99" "g100" "g107" "g110" "g122" "g124" "g125" "g129" "g134" "g139"
## [40] "g141" "g143" "g144" "g146" "g148" "g154" "g163" "g165" "g171" "g173" "g178" "g180" "g182"
## [53] "g188" "g190" "g193" "g195"
##
## $t4
## [1] "g2" "g5" "g7" "g8" "g9" "g12" "g13" "g15" "g20" "g21" "g26" "g28" "g30"
## [14] "g31" "g36" "g37" "g38" "g47" "g63" "g64" "g65" "g67" "g68" "g69" "g76" "g77"
## [27] "g80" "g85" "g90" "g98" "g99" "g101" "g105" "g106" "g120" "g123" "g125" "g126" "g129"
## [40] "g131" "g134" "g137" "g140" "g143" "g147" "g148" "g153" "g155" "g157" "g165" "g167" "g170"
## [53] "g171" "g172" "g174" "g175" "g176" "g178" "g181" "g182" "g188" "g192" "g199"
- Return all row labels (genes) that match the above query across a specified number of columns
(here 2). Note, the
rowSumsfunction is used for this, which performs the row-wise looping internally and extremely fast.
matchingID <- rowSums(queryResult) > 2
queryResult[matchingID, , drop=FALSE]
## t1 t2 t3 t4
## g5 TRUE TRUE FALSE TRUE
## g7 FALSE TRUE TRUE TRUE
## g9 FALSE TRUE TRUE TRUE
## g12 FALSE TRUE TRUE TRUE
## g23 TRUE TRUE TRUE FALSE
## g80 FALSE TRUE TRUE TRUE
## g85 FALSE TRUE TRUE TRUE
## g88 TRUE TRUE TRUE FALSE
## g90 FALSE TRUE TRUE TRUE
## g99 FALSE TRUE TRUE TRUE
## g120 TRUE TRUE FALSE TRUE
## g129 FALSE TRUE TRUE TRUE
## g134 TRUE FALSE TRUE TRUE
## g148 FALSE TRUE TRUE TRUE
## g165 FALSE TRUE TRUE TRUE
## g170 TRUE TRUE FALSE TRUE
## g171 TRUE FALSE TRUE TRUE
## g172 TRUE TRUE FALSE TRUE
## g178 TRUE TRUE TRUE TRUE
names(matchingID[matchingID])
## [1] "g5" "g7" "g9" "g12" "g23" "g80" "g85" "g88" "g90" "g99" "g120" "g129" "g134"
## [14] "g148" "g165" "g170" "g171" "g172" "g178"
As demonstrated in the above query examples, by setting up the proper data structures (here two
matrices with same dimensions), and utilizing vectorized (matrix-to-matrix) operations
along with R’s built-in row* and col* stats function family (e.g. rowSums) one can
design with very little code flexible query routines that also run very time-efficient.
Functions
Function Overview
A very useful feature of the R environment is the possibility to expand existing functions and to easily write custom functions. In fact, most of the R software can be viewed as a series of R functions.
Syntax to define function
myfct <- function(arg1, arg2, ...) {
function_body
}
Syntax to call functions
myfct(arg1=..., arg2=...)
The value returned by a function is the value of the function body, which is usually an unassigned final expression, e.g.: return()
Function Syntax Rules
General
- Functions are defined by
- The assignment with the keyword
function - The declaration of arguments/variables (
arg1, arg2, ...) - The definition of operations (
function_body) that perform computations on the provided arguments. A function name needs to be assigned to call the function.
- The assignment with the keyword
Naming
- Function names can be almost anything. However, the usage of names of existing functions should be avoided.
Arguments
- It is often useful to provide default values for arguments (e.g.:
arg1=1:10). This way they don’t need to be provided in a function call. The argument list can also be left empty (myfct <- function() { fct_body }) if a function is expected to return always the same value(s). The argument...can be used to allow one function to pass on argument settings to another.
Body
- The actual expressions (commands/operations) are defined in the function body which should be enclosed by braces. The individual commands are separated by semicolons or new lines (preferred).
Usage
- Functions are called by their name followed by parentheses containing possible argument names. Empty parenthesis after the function name will result in an error message when a function requires certain arguments to be provided by the user. The function name alone will print the definition of a function.
Scope
- Variables created inside a function exist only for the life time of a function. Thus, they are not accessible outside of the function. To force variables in functions to exist globally, one can use the double assignment operator:
<<-
Examples
Define sample function
myfct <- function(x1, x2=5) {
z1 <- x1 / x1
z2 <- x2 * x2
myvec <- c(z1, z2)
return(myvec)
}
Function usage
Apply function to values 2 and 5
myfct(x1=2, x2=5)
## [1] 1 25
Run without argument names
myfct(2, 5)
## [1] 1 25
Makes use of default value 5
myfct(x1=2)
## [1] 1 25
Print function definition (often unintended)
myfct
## function(x1, x2=5) {
## z1 <- x1 / x1
## z2 <- x2 * x2
## myvec <- c(z1, z2)
## return(myvec)
## }
## <bytecode: 0x56dd086c6988>
Programming Exercise on Functions: go here https://girke.bioinformatics.ucr.edu/GEN242/tutorials/rprogramming/rprogramming/#exercise-1)
Useful Utilities
Debugging Utilities
Several debugging utilities are available for R. They include:
tracebackbrowseroptions(error=recover)options(error=NULL)debug
The Debugging in R page provides an overview of the available resources.
Regular Expressions
R’s regular expression utilities work similar as in other languages. To learn how to use them in R, one can consult the main help page on this topic with ?regexp.
String matching with grep
The grep function can be used for finding patterns in strings, here letter A in vector month.name.
month.name[grep("A", month.name)]
## [1] "April" "August"
String substitution with gsub
Example for using regular expressions to substitute a pattern by another one using a back reference. Remember: single escapes \ need to be double escaped \\ in R.
gsub('(i.*a)', 'xxx_\\1', "virginica", perl = TRUE)
## [1] "vxxx_irginica"
Interpreting a Character String as Expression
Some useful examples
Generates vector of object names in session
myfct <- function(x) x^2
mylist <- ls()
n <- which(mylist %in% "myfct")
mylist[n]
## [1] "myfct"
Executes entry in position n as expression
get(mylist[n])
## function(x) x^2
get(mylist[n])(2)
## [1] 4
Alternative approach
eval(parse(text=mylist[n]))
## function(x) x^2
Replacement, Split and Paste Functions for Strings
Selected examples
Substitution with back reference which inserts in this example _ character
x <- gsub("(a)","\\1_", month.name[1], perl=T)
x
## [1] "Ja_nua_ry"
Split string on inserted character from above
strsplit(x,"_")
## [[1]]
## [1] "Ja" "nua" "ry"
Reverse a character string by splitting first all characters into vector fields
paste(rev(unlist(strsplit(x, NULL))), collapse="")
## [1] "yr_aun_aJ"
Check Integrity of Files
The integrity of files (e.g. after downloading or copying them) can be checked
with md5sum (also see checkMD5sums).
library(tools)
(md5 <- as.vector(md5sum(dir(R.home(), pattern = "^COPY", full.names = TRUE))))
## [1] "b234ee4d69f5fce4486a80fdaf4a4263"
identical(md5, md5)
## [1] TRUE
identical(md5, sub("^b", "z", md5))
## [1] FALSE
Time, Date and Sleep
Selected examples
Return CPU (and other) times that an expression used (here ls)
system.time(ls())
## user system elapsed
## 0 0 0
Return the current system date and time
date()
## [1] "Thu Apr 14 09:21:47 2022"
Pause execution of R expressions for a given number of seconds (e.g. in loop)
Sys.sleep(1)
Example
Import of Specific File Lines with Regular Expression
The following example demonstrates the retrieval of specific lines from an external file with a regular expression. First, an external file is created with the cat function, all lines of this file are imported into a vector with readLines, the specific elements (lines) are then retieved with the grep function, and the resulting lines are split into vector fields with strsplit.
cat(month.name, file="zzz.txt", sep="\n")
x <- readLines("zzz.txt")
x[1:6]
## [1] "January" "February" "March" "April" "May" "June"
x <- x[c(grep("^J", as.character(x), perl = TRUE))]
t(as.data.frame(strsplit(x, "u")))
## [,1] [,2]
## c..Jan....ary.. "Jan" "ary"
## c..J....ne.. "J" "ne"
## c..J....ly.. "J" "ly"
Calling External Software
External command-line software can be called with the system and system2 functions. The following example calls blastall from R.
system("blastall -p blastp -i seq.fasta -d uniprot -o seq.blastp")
Possibilities for Executing R Scripts
R console
The source command is used for executing an R script from the R console.
source("my_script.R")
Command-line
The Rscript commmand is used to execute an R script from the command-line.
Rscript myscript.R
./myscript.R # also possible after including a shebang in myscript.R and making it executable
A shebang needs to be included at the beginning of an R script in order to execute it just by typing the name of the script (here ./myscript.R) after making it executable (see here for UNIX-like OSs). The shebang line is optional when executing R scripts from the R console with the source command or with Rscript from the command-line. The shebang of R scripts looks as follows:
#!/usr/bin/env Rscript
Older alternatives for executing R scripts from the command-line are given below.
R CMD BATCH myscript.R # Alternative way 1
R --slave < myscript.R # Alternative way 2
Passing arguments from command-line to R
Create an R script named test.R with the following content:
myarg <- commandArgs()
print(iris[1:myarg[6], ])
Then run it from the command-line like this:
Rscript test.R 10
In the given example the number 10 is passed on from the command-line as an argument to the R script which is used to return to STDOUT the first 10 rows of the iris sample data. If several arguments are provided, they will be interpreted as one string and need to be split in R with the strsplit function. A more detailed example can be found here.
Building R Packages
This section has been moved to a dedicated tutorial on R package development here.
Programming Exercises
Exercise 1
for loop
Task 1.1: Compute the mean of each row in myMA by applying the mean function in a for loop.
myMA <- matrix(rnorm(500), 100, 5, dimnames=list(1:100, paste("C", 1:5, sep="")))
myve_for <- NULL
for(i in seq(along=myMA[,1])) {
myve_for <- c(myve_for, mean(as.numeric(myMA[i, ])))
}
myResult <- cbind(myMA, mean_for=myve_for)
myResult[1:4, ]
## C1 C2 C3 C4 C5 mean_for
## 1 -1.202619 -0.1053735 -1.1074995 0.4269033 -1.3932608 -0.67636998
## 2 1.174138 -0.8991985 -1.2658862 1.0151751 -0.4075398 -0.07666225
## 3 1.357860 -1.2370991 0.7402124 -1.0277818 0.4851062 0.06365953
## 4 1.173512 -1.2724114 0.2270477 -1.5421106 0.7484669 -0.13309912
while loop
Task 1.2: Compute the mean of each row in myMA by applying the mean function in a while loop.
z <- 1
myve_while <- NULL
while(z <= length(myMA[,1])) {
myve_while <- c(myve_while, mean(as.numeric(myMA[z, ])))
z <- z + 1
}
myResult <- cbind(myMA, mean_for=myve_for, mean_while=myve_while)
myResult[1:4, -c(1,2)]
## C3 C4 C5 mean_for mean_while
## 1 -1.1074995 0.4269033 -1.3932608 -0.67636998 -0.67636998
## 2 -1.2658862 1.0151751 -0.4075398 -0.07666225 -0.07666225
## 3 0.7402124 -1.0277818 0.4851062 0.06365953 0.06365953
## 4 0.2270477 -1.5421106 0.7484669 -0.13309912 -0.13309912
Task 1.3: Confirm that the results from both mean calculations are identical
all(myResult[,6] == myResult[,7])
## [1] TRUE
apply loop
Task 1.4: Compute the mean of each row in myMA by applying the mean function in an apply loop
myve_apply <- apply(myMA, 1, mean)
myResult <- cbind(myMA, mean_for=myve_for, mean_while=myve_while, mean_apply=myve_apply)
myResult[1:4, -c(1,2)]
## C3 C4 C5 mean_for mean_while mean_apply
## 1 -1.1074995 0.4269033 -1.3932608 -0.67636998 -0.67636998 -0.67636998
## 2 -1.2658862 1.0151751 -0.4075398 -0.07666225 -0.07666225 -0.07666225
## 3 0.7402124 -1.0277818 0.4851062 0.06365953 0.06365953 0.06365953
## 4 0.2270477 -1.5421106 0.7484669 -0.13309912 -0.13309912 -0.13309912
Avoiding loops
Task 1.5: When operating on large data sets it is much faster to use the rowMeans function
mymean <- rowMeans(myMA)
myResult <- cbind(myMA, mean_for=myve_for, mean_while=myve_while, mean_apply=myve_apply, mean_int=mymean)
myResult[1:4, -c(1,2,3)]
## C4 C5 mean_for mean_while mean_apply mean_int
## 1 0.4269033 -1.3932608 -0.67636998 -0.67636998 -0.67636998 -0.67636998
## 2 1.0151751 -0.4075398 -0.07666225 -0.07666225 -0.07666225 -0.07666225
## 3 -1.0277818 0.4851062 0.06365953 0.06365953 0.06365953 0.06365953
## 4 -1.5421106 0.7484669 -0.13309912 -0.13309912 -0.13309912 -0.13309912
To find out which other built-in functions for basic calculations exist, type ?rowMeans.
Exercise 2
Custom functions
Task 2.1: Use the following code as basis to implement a function that allows the user to compute the mean for any combination of columns in a matrix or data frame. The first argument of this function should specify the input data set, the second the mathematical function to be passed on (e.g. mean, sd, max) and the third one should allow the selection of the columns by providing a grouping vector.
myMA <- matrix(rnorm(100000), 10000, 10, dimnames=list(1:10000, paste("C", 1:10, sep="")))
myMA[1:2,]
## C1 C2 C3 C4 C5 C6 C7 C8
## 1 0.1750576 -0.2826930 0.9290137 0.0830523803 1.4113331 -1.221849 -0.4663186 0.6170635
## 2 -0.8056727 0.1885557 0.4891758 -0.0003536156 0.8356054 1.406484 1.0363437 -0.1351562
## C9 C10
## 1 0.58305534 0.1716817
## 2 0.09596605 0.4551591
myList <- tapply(colnames(myMA), c(1,1,1,2,2,2,3,3,4,4), list)
names(myList) <- sapply(myList, paste, collapse="_")
myMAmean <- sapply(myList, function(x) apply(myMA[,x], 1, mean))
myMAmean[1:4,]
## C1_C2_C3 C4_C5_C6 C7_C8 C9_C10
## 1 0.27379276 0.0908453513 0.07537248 0.3773685
## 2 -0.04264706 0.7472451361 0.45059378 0.2755626
## 3 0.49411343 -0.4486259202 -0.08519687 1.0929435
## 4 0.06575562 -0.0007494971 0.07968924 -0.7374505
Exercise 3
Nested loops to generate similarity matrices
Task 3.1: Create a sample list populated with character vectors of different lengths
setlist <- lapply(11:30, function(x) sample(letters, x, replace=TRUE))
names(setlist) <- paste("S", seq(along=setlist), sep="")
setlist[1:6]
## $S1
## [1] "b" "s" "o" "w" "w" "x" "s" "z" "d" "f" "b"
##
## $S2
## [1] "e" "w" "l" "y" "i" "l" "m" "q" "s" "r" "d" "s"
##
## $S3
## [1] "h" "i" "t" "d" "f" "c" "u" "r" "v" "a" "n" "t" "v"
##
## $S4
## [1] "o" "w" "c" "x" "c" "l" "a" "z" "c" "f" "n" "i" "j" "u"
##
## $S5
## [1] "a" "z" "b" "t" "p" "l" "u" "z" "q" "x" "y" "h" "l" "j" "e"
##
## $S6
## [1] "z" "v" "n" "v" "b" "m" "p" "p" "z" "g" "l" "j" "z" "z" "t" "e"
Task 3.2: Compute the length for all pairwise intersects of the vectors stored in setlist. The intersects can be determined with the %in% function like this: sum(setlist[[1]] %in% setlist[[2]])
setlist <- sapply(setlist, unique)
olMA <- sapply(names(setlist), function(x) sapply(names(setlist),
function(y) sum(setlist[[x]] %in% setlist[[y]])))
olMA[1:12,]
## S1 S2 S3 S4 S5 S6 S7 S8 S9 S10 S11 S12 S13 S14 S15 S16 S17 S18 S19 S20
## S1 8 3 2 5 3 2 4 4 4 5 8 5 5 5 6 5 4 6 4 4
## S2 3 10 3 3 4 3 5 3 3 5 4 6 6 8 7 5 5 8 7 8
## S3 2 3 11 6 4 3 5 4 4 7 4 8 4 5 8 6 6 6 7 8
## S4 5 3 6 12 6 4 8 4 4 8 8 7 7 7 8 9 6 8 9 9
## S5 3 4 4 6 13 7 3 6 6 8 7 8 5 10 8 9 6 10 10 10
## S6 2 3 3 4 7 11 4 5 5 5 5 7 4 8 7 6 8 8 9 8
## S7 4 5 5 8 3 4 11 5 6 5 6 6 7 6 9 6 7 7 8 9
## S8 4 3 4 4 6 5 5 12 7 4 7 6 5 7 10 7 8 8 9 7
## S9 4 3 4 4 6 5 6 7 11 6 8 6 3 8 10 6 6 8 8 8
## S10 5 5 7 8 8 5 5 4 6 14 7 9 5 9 9 10 7 11 9 8
## S11 8 4 4 8 7 5 6 7 8 7 13 7 6 10 10 9 8 9 8 8
## S12 5 6 8 7 8 7 6 6 6 9 7 15 5 10 10 9 9 11 10 10
Task 3.3 Plot the resulting intersect matrix as heat map.
The image or the pheatmap functions can be used for this.
library(pheatmap); library("RColorBrewer")
pheatmap(olMA, color=brewer.pal(9,"Blues"), cluster_rows=FALSE, cluster_cols=FALSE, display_numbers=TRUE, number_format="%.0f", fontsize_number=10)
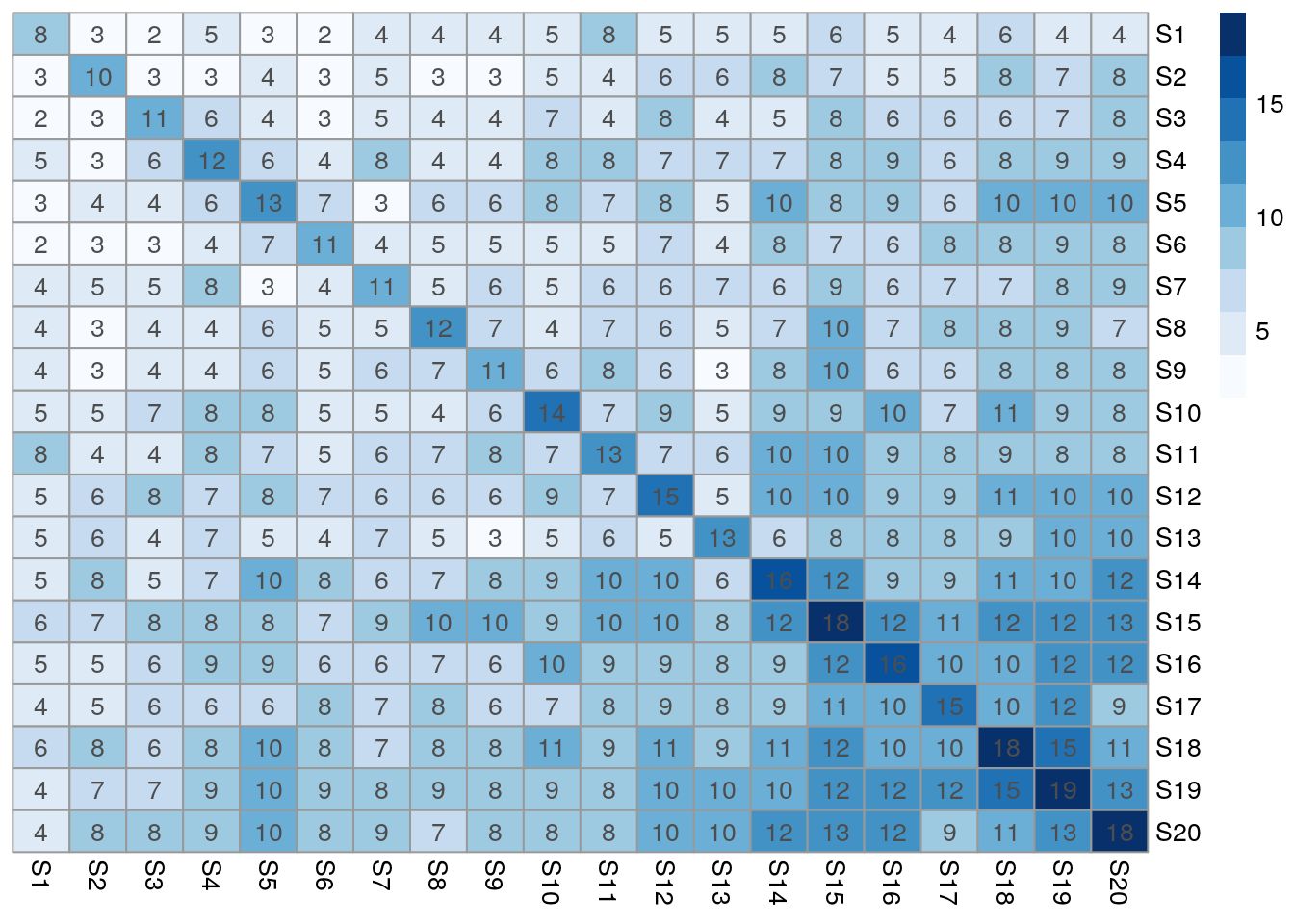
# image(olMA)
Exercise 4
Build your own R package
Task 4.1: Save one or more of your functions to a file called script.R and build the package with the package.skeleton function.
package.skeleton(name="mypackage", code_files=c("script1.R"))
Task 4.2: Build tarball of the package
system("R CMD build mypackage")
Task 4.3: Install and use package
install.packages("mypackage_1.0.tar.gz", repos=NULL, type="source")
library(mypackage)
?myMAcomp # Opens help for function defined by mypackage
Homework 5
See homework section here.
Additional Exercises
Pattern matching and positional parsing of equences
The following sample script patternSearch.R defines functions for importing sequences into R, retrieving reverse and complement of nucleotide sequences, pattern searching, positional parsing and exporting search results in HTML format. Sourcing the script will return usage instructions of its functions.
source("https://raw.githubusercontent.com/tgirke/GEN242/main/content/en/tutorials/rprogramming/scripts/patternSearch.R")
Identify over-represented strings in sequence sets
Example functions for finding over-represented words in sets of DNA, RNA or protein sequences are defined in this script: wordFinder.R. Sourcing the script will return usage instructions of its functions.
source("https://raw.githubusercontent.com/tgirke/GEN242/main/content/en/tutorials/rprogramming/scripts/wordFinder.R")
Object-Oriented Programming (OOP)
R supports several systems for object-oriented programming (OOP). This includes an older S3 system, and the more recently introduced R6 and S4 systems. The latter is the most formal version that supports multiple inheritance, multiple dispatch and introspection. Many of these features are not available in the older S3 system. In general, the OOP approach taken by R is to separate the class specifications from the specifications of generic functions (function-centric system). The following introduction is restricted to the S4 system since it is nowadays the preferred OOP method for package development in Bioconductor. More information about OOP in R can be found in the following introductions:
- Vincent Zoonekynd’s introduction to S3 Classes
- Christophe Genolini’s S4 Intro
- Advanced Bioconductor Courses
- Programming with R by John Chambers
- R Programming for Bioinformatics by Robert Gentleman
- Advanced R online book by Hadley Wichham
Define S4 Classes
1. Define S4 Classes with setClass() and new()
y <- matrix(1:10, 2, 5) # Sample data set
setClass(Class="myclass",
representation=representation(a="ANY"),
prototype=prototype(a=y[1:2,]), # Defines default value (optional)
validity=function(object) { # Can be defined in a separate step using setValidity
if(class(object@a)[1]!="matrix") {
return(paste("expected matrix, but obtained", class(object@a)))
} else {
return(TRUE)
}
}
)
The setClass function defines classes. Its most important arguments are
Class: the name of the classrepresentation: the slots that the new class should have and/or other classes that this class extends.prototype: an object providing default data for the slots.contains: the classes that this class extends.validity,access,version: control arguments included for compatibility with S-Plus.where: the environment to use to store or remove the definition as meta data.
2. Create new class instance
The function new creates an instance of a class (here myclass).
myobj <- new("myclass", a=y)
myobj
## An object of class "myclass"
## Slot "a":
## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 3 5 7 9
## [2,] 2 4 6 8 10
If evaluated the following would return an error due to wrong input type (data.frame instead of matrix).
new("myclass", a=iris) # Returns error due to wrong input
The arguments of new are:
Class: the name of the class...: data to include in the new object with arguments according to slots in class definition
3. Initialization method
A more generic way of creating class instances is to define an initialization method (more details below).
setMethod("initialize", "myclass", function(.Object, a) {
.Object@a <- a/a
.Object
})
new("myclass", a = y)
## An object of class "myclass"
## Slot "a":
## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 1 1 1 1
## [2,] 1 1 1 1 1
4. Usage and helper functions
The ‘@’ operator extracts the contents of a slot. Its usage should be limited to internal functions.
myobj@a
## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 3 5 7 9
## [2,] 2 4 6 8 10
Create a new S4 object from an old one.
initialize(.Object=myobj, a=as.matrix(cars[1:2,]))
## An object of class "myclass"
## Slot "a":
## speed dist
## 1 1 1
## 2 1 1
If evaluated the removeClass function removes an object from the current session.
This does not apply to associated methods.
removeClass("myclass")
5. Inheritance
Inheritance allows to define new classes that inherit all properties (e.g. data slots, methods)
from their existing parent classes. The contains argument used below allows to extend
existing classes. This propagates all slots of parent classes.
setClass("myclass1", representation(a = "character", b = "character"))
setClass("myclass2", representation(c = "numeric", d = "numeric"))
setClass("myclass3", contains=c("myclass1", "myclass2"))
new("myclass3", a=letters[1:4], b=letters[1:4], c=1:4, d=4:1)
## An object of class "myclass3"
## Slot "a":
## [1] "a" "b" "c" "d"
##
## Slot "b":
## [1] "a" "b" "c" "d"
##
## Slot "c":
## [1] 1 2 3 4
##
## Slot "d":
## [1] 4 3 2 1
getClass("myclass1")
## Class "myclass1" [in ".GlobalEnv"]
##
## Slots:
##
## Name: a b
## Class: character character
##
## Known Subclasses: "myclass3"
getClass("myclass2")
## Class "myclass2" [in ".GlobalEnv"]
##
## Slots:
##
## Name: c d
## Class: numeric numeric
##
## Known Subclasses: "myclass3"
getClass("myclass3")
## Class "myclass3" [in ".GlobalEnv"]
##
## Slots:
##
## Name: a b c d
## Class: character character numeric numeric
##
## Extends: "myclass1", "myclass2"
6. Coerce objects to another class
The following defines a coerce method. After this the standard as(..., "...")
syntax can be used to coerce the new class to another one.
setAs(from="myclass", to="character", def=function(from) as.character(as.matrix(from@a)))
as(myobj, "character")
## [1] "1" "2" "3" "4" "5" "6" "7" "8" "9" "10"
7. Virtual classes
Virtual classes are constructs for which no instances will be or can be
created. They are used to link together classes which may have distinct
representations (e.g. cannot inherit from each other) but for which one wants
to provide similar functionality. Often it is desired to create a virtual class
and to then have several other classes extend it. Virtual classes can be
defined by leaving out the representation argument or including the class
VIRTUAL as illustrated here:
setClass("myVclass")
setClass("myVclass", representation(a = "character", "VIRTUAL"))
8. Introspection of classes
Useful functions to introspect classes include:
getClass("myclass")getSlots("myclass")slotNames("myclass")extends("myclass2")
Assign Generics and Methods
Generics and methods can be assigned with the methods setGeneric() and setMethod().
1. Accessor functions
This avoids the usage of the @ operator.
setGeneric(name="acc", def=function(x) standardGeneric("acc"))
## [1] "acc"
setMethod(f="acc", signature="myclass", definition=function(x) {
return(x@a)
})
acc(myobj)
## [,1] [,2] [,3] [,4] [,5]
## [1,] 1 3 5 7 9
## [2,] 2 4 6 8 10
2. Replacement methods
- Using custom accessor function with
acc <-syntax.
setGeneric(name="acc<-", def=function(x, value) standardGeneric("acc<-"))
## [1] "acc<-"
setReplaceMethod(f="acc", signature="myclass", definition=function(x, value) {
x@a <- value
return(x)
})
## After this the following replace operations with 'acc' work on new object class
acc(myobj)[1,1] <- 999 # Replaces first value
colnames(acc(myobj)) <- letters[1:5] # Assigns new column names
rownames(acc(myobj)) <- letters[1:2] # Assigns new row names
myobj
## An object of class "myclass"
## Slot "a":
## a b c d e
## a 999 3 5 7 9
## b 2 4 6 8 10
- Replacement method using
[operator, here[...] <-syntax.
setReplaceMethod(f="[", signature="myclass", definition=function(x, i, j, value) {
x@a[i,j] <- value
return(x)
})
myobj[1,2] <- 999
myobj
## An object of class "myclass"
## Slot "a":
## a b c d e
## a 999 999 5 7 9
## b 2 4 6 8 10
3. Behavior of bracket operator
The behavior of the bracket [ subsetting operator can be defined as follows.
setMethod(f="[", signature="myclass",
definition=function(x, i, j, ..., drop) {
x@a <- x@a[i,j]
return(x)
})
myobj[1:2, 1:3] # Standard subsetting works now on new class
## An object of class "myclass"
## Slot "a":
## a b c
## a 999 999 5
## b 2 4 6
4. Print behavior
A convient summary printing behavior for a new class should always be defined.
setMethod(f="show", signature="myclass", definition=function(object) {
cat("An instance of ", "\"", class(object), "\"", " with ", length(acc(object)[,1]), " elements", "\n", sep="")
if(length(acc(object)[,1])>=5) {
print(as.data.frame(rbind(acc(object)[1:2,], ...=rep("...", length(acc(object)[1,])), acc(object)[(length(acc(object)[,1])-1):length(acc(object)[,1]),])))
} else {
print(acc(object))
}
})
myobj # Prints object with custom method
## An instance of "myclass" with 2 elements
## a b c d e
## a 999 999 5 7 9
## b 2 4 6 8 10
5. Define custom methods
The following gives an example for defining a data specific method, here randomizing row order of matrix stored in new S4 class.
setGeneric(name="randomize", def=function(x) standardGeneric("randomize"))
## [1] "randomize"
setMethod(f="randomize", signature="myclass", definition=function(x) {
acc(x)[sample(1:length(acc(x)[,1]), length(acc(x)[,1])), ]
})
randomize(myobj)
## a b c d e
## b 2 4 6 8 10
## a 999 999 5 7 9
6. Plotting method
Define a graphical plotting method for new class and allow users to access it with
R’s generic plot function.
setMethod(f="plot", signature="myclass", definition=function(x, ...) {
barplot(as.matrix(acc(x)), ...)
})
plot(myobj)
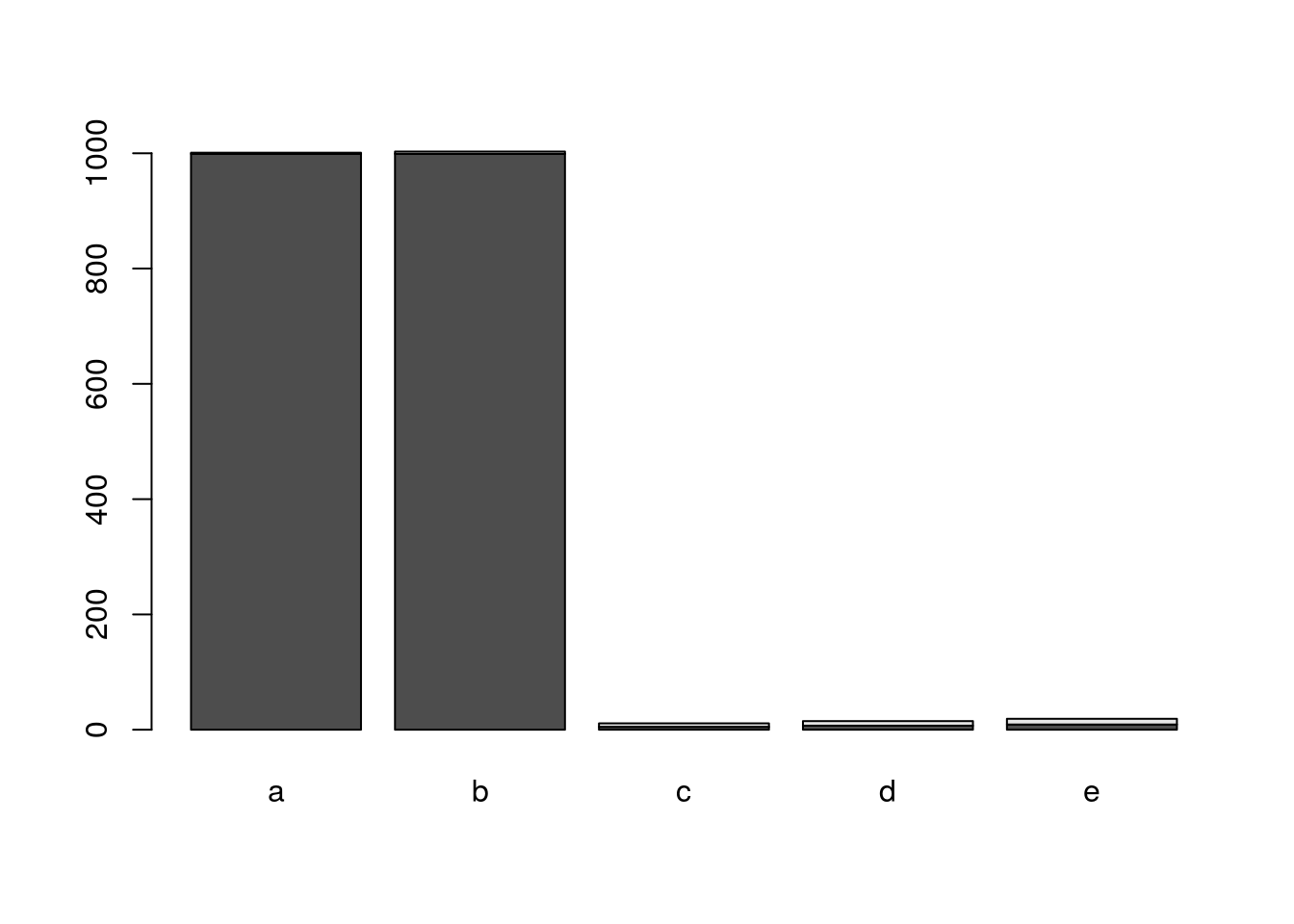
7. Utilities to inspect methods
Important inspection methods for classes include:
showMethods(class="myclass")findMethods("randomize")getMethod("randomize", signature="myclass")existsMethod("randomize", signature="myclass")
Session Info
sessionInfo()
## R version 4.3.0 (2023-04-21)
## Platform: x86_64-pc-linux-gnu (64-bit)
## Running under: Debian GNU/Linux 11 (bullseye)
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8
## [4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C
## [10] LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: America/Los_Angeles
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] RColorBrewer_1.1-3 pheatmap_1.0.12 BiocStyle_2.28.0
##
## loaded via a namespace (and not attached):
## [1] cli_3.6.1 knitr_1.42 rlang_1.1.1 xfun_0.39
## [5] jsonlite_1.8.4 glue_1.6.2 colorspace_2.1-0 htmltools_0.5.5
## [9] sass_0.4.5 scales_1.2.1 rmarkdown_2.21 grid_4.3.0
## [13] munsell_0.5.0 evaluate_0.20 jquerylib_0.1.4 fastmap_1.1.1
## [17] yaml_2.3.7 lifecycle_1.0.3 bookdown_0.33 BiocManager_1.30.20
## [21] compiler_4.3.0 blogdown_1.16 digest_0.6.31 R6_2.5.1
## [25] bslib_0.4.2 tools_4.3.0 gtable_0.3.3 cachem_1.0.7
References
Gentleman, Robert. 2008. R Programming for Bioinformatics (Chapman & Hall/CRC Computer Science & Data Analysis). 1 edition. Chapman; Hall/CRC. http://www.amazon.com/Programming-Bioinformatics-Chapman-Computer-Analysis/dp/1420063677.