Automate Creation of CWL Instructions
16 minute read
Introduction
A central concept for designing workflows within the systemPipeR environment
is the usage of workflow management containers. For describing analysis
workflows in a generic and flexible manner the Common Workflow
Language (CWL) has been adopted throughout the
environment including the workflow management containers (Amstutz et al. 2016).
Using the CWL community standard in systemPipeR has many advantages. For
instance, the integration of CWL allows running systemPipeR workflows from a
single specification instance either entirely from within R, from various
command line wrappers (e.g., cwl-runner) or from other languages (e.g., Bash or
Python). An important feature of systemPipeR's CWL interface is that it
provides two options to run command line tools and workflows based on CWL.
First, one can run CWL in its native way via an R-based wrapper utility for
cwl-runner or cwl-tools (CWL-based approach). Second, one can run workflows
using CWL’s command line and workflow instructions from within R (R-based
approach). In the latter case the same CWL workflow definition files (e.g.
.cwl and .yml) are used but rendered and executed entirely with R functions
defined by systemPipeR, and thus use CWL mainly as a command line and
workflow definition format rather than execution software to run workflows.
Moreover, systemPipeR provides several convenience functions that are useful
for designing and debugging workflows, such as a command-line rendering
function to retrieve the exact command-line strings for each data set and
processing step prior to running a command-line.
This tutorial briefly introduces the basics how CWL defines command-line
syntax. Next, it describes how to use CWL within systemPipeR for designing,
modifying and running workflows.
Load package
Recent versions of R (>=4.0.0), Bioconductor (>=3.14) and systemPipeR (>=2.0.8)
need to be used to gain access to the functions described in this tutorial.
CWL command line specifications
CWL command line specifications are written in YAML format.
In CWL, files with the extension .cwl define the parameters of a chosen
command line step or workflow, while files with the extension .yml define
the input variables of command line steps.
The following introduces first the basic structure of .cwl files.
dir_path <- system.file("extdata/cwl/example/", package="systemPipeR")
cwl <- yaml::read_yaml(file.path(dir_path, "example.cwl"))
- The
cwlVersioncomponent specifies the version of CWL that is used here. - The
classcomponent declares the usage of a command-line tool. Note, CWL has anotherclasscalledWorkflow. The latter defines one or more command-line tools, whileCommandLineToolis limited to one.
cwl[1:2]
## $cwlVersion
## [1] "v1.0"
##
## $class
## [1] "CommandLineTool"
- The
baseCommandcomponent contains the base name of the software to be executed.
cwl[3]
## $baseCommand
## [1] "echo"
- The
inputscomponent provides the input information required for the command-line software. Important sub-components of this section are:id: each input has an id assigning a nametype: input type value (e.g. string, int, long, float, double, File, Directory or Any);inputBinding: optional component indicating if the input parameter should appear on the command line. If missing then the parameter will not appear in the command-line.
cwl[4]
## $inputs
## $inputs$message
## $inputs$message$type
## [1] "string"
##
## $inputs$message$inputBinding
## $inputs$message$inputBinding$position
## [1] 1
##
##
##
## $inputs$SampleName
## $inputs$SampleName$type
## [1] "string"
##
##
## $inputs$results_path
## $inputs$results_path$type
## [1] "Directory"
- The
outputscomponent should provide a list of the outputs expected after running a command-line tools. Important sub-components of this section are:id: each output has an id assigning a nametype: output type value (e.g. string, int, long, float, double, File, Directory, Any orstdout)outputBinding: defines how to set the outputs values. Theglobcomponent will define the name of the output value.
cwl[5]
## $outputs
## $outputs$string
## $outputs$string$type
## [1] "stdout"
stdout: specifies afilenamefor capturing standard output. Note here we are using a syntax that takes advantage of the inputs section, usingresults_pathparameter and also theSampleNameto construct thefilenameof the output.
cwl[6]
## $stdout
## [1] "$(inputs.results_path.basename)/$(inputs.SampleName).txt"
Next, the structure and content of the .yml files will be introduced. The .yml file
provides the parameter values for the .cwl components described above.
The following example defines three parameters.
yaml::read_yaml(file.path(dir_path, "example_single.yml"))
## $message
## [1] "Hello World!"
##
## $SampleName
## [1] "M1"
##
## $results_path
## $results_path$class
## [1] "Directory"
##
## $results_path$path
## [1] "./results"
Importantly, if an input component is defined in the corresponding .cwl file, then the required value needs to be provided by the corresponding component of the .yml file.
How to connect CWL description files within systemPipeR
A SYSargsList container stores several SYSargs2 instances in a list-like object containing
all instructions required for processing a set of input files with a single or many command-line
steps within a workflow (i.e. several tools of one software or several independent software tools).
A single SYSargs2 object is created and fully populated with the constructor functions
loadWF and renderWF.
The following imports a .cwl file (here example.cwl) for running a simple echo Hello World
example where a string Hello World will be printed to stdout and redirected to a file named
M1.txt located under a subdirectory named results.
HW <- loadWF(wf_file="example.cwl", input_file="example_single.yml",
dir_path = dir_path)
HW <- renderWF(HW)
HW
## Instance of 'SYSargs2':
## Slot names/accessors:
## targets: 0 (...), targetsheader: 0 (lines)
## modules: 0
## wf: 0, clt: 1, yamlinput: 3 (inputs)
## input: 1, output: 1
## cmdlist: 1
## Sub Steps:
## 1. example (rendered: TRUE)
cmdlist(HW)
## $defaultid
## $defaultid$example
## [1] "echo Hello World! > results/M1.txt"
The above example is limited to running only one command-line call, corresponding to one
input file, e.g. representing a single experimental sample. To scale to many command-line
calls, e.g. when processing many input samples, a simple solution offered by systemPipeR
is to use variables, one for each parameter with many inputs.
The following gives a simple example for defining and processing many inputs.
yml <- yaml::read_yaml(file.path(dir_path, "example.yml"))
yml
## $message
## [1] "_STRING_"
##
## $SampleName
## [1] "_SAMPLE_"
##
## $results_path
## $results_path$class
## [1] "Directory"
##
## $results_path$path
## [1] "./results"
Under the message and SampleName parameters, variables are used for that will be populated
by values provided by a third file called targets.
The following shows the structure of a simple targets file.
targetspath <- system.file("extdata/cwl/example/targets_example.txt", package="systemPipeR")
read.delim(targetspath, comment.char = "#")
## Message SampleName
## 1 Hello World! M1
## 2 Hello USA! M2
## 3 Hello Bioconductor! M3
With help of a targets file, one can define all input files, sample ids and
experimental variables relevant for an analysis workflow. In the above example,
strings defined under the Message column will be passed on to the echo
command-line tool. In addition, each command-line will be assigned a label or
id specified under SampleName column. Any number of additional columns can be
added as needed.
Users should note here, the usage of targets files is optional when using
systemPipeR's CWL interface. Since targets files are very efficient for
organizing experimental variables, their usage is highly encouraged and well
supported in systemPipeR.
Connect parameter and targets files
The constructor functions construct an SYSargs2 instance from three input files:
- `.cwl` file path assigned to `wf_file` argument
- `.yml` file path assigned to `input_file` argument
- `target` file assigned to `targets` argument
As mentioned above, the latter targets file is optional. The connection
between input variables (here defined by input_file argument) and the
targets file are defined under the inputvars argument. A named vector is
required, where each element name needs to match the column names in the
targets file, and the value must match the names of the .yml variables.
This is used to replace the CWL variable and construct the command-lines, usually
one for each input sample.
For consistency the pattern _XXXX_ is used for variable naming in the .yml file, where the
name matches the corresponding column name in the targets file. This pattern is recommended
for easy identification but not enforced.
The following imports a .cwl file (same example as above) for running
the echo example. However, now several command-line calls are constructed with the
information provided under the Message column of the targets file that is passed on to
matching component in the .yml file.
HW_mul <- loadWorkflow(targets = targetspath, wf_file="example.cwl",
input_file="example.yml", dir_path = dir_path)
HW_mul <- renderWF(HW_mul, inputvars = c(Message = "_STRING_", SampleName = "_SAMPLE_"))
HW_mul
## Instance of 'SYSargs2':
## Slot names/accessors:
## targets: 3 (M1...M3), targetsheader: 1 (lines)
## modules: 0
## wf: 0, clt: 1, yamlinput: 3 (inputs)
## input: 3, output: 3
## cmdlist: 3
## Sub Steps:
## 1. example (rendered: TRUE)
cmdlist(HW_mul)
## $M1
## $M1$example
## [1] "echo Hello World! > results/M1.txt"
##
##
## $M2
## $M2$example
## [1] "echo Hello USA! > results/M2.txt"
##
##
## $M3
## $M3$example
## [1] "echo Hello Bioconductor! > results/M3.txt"
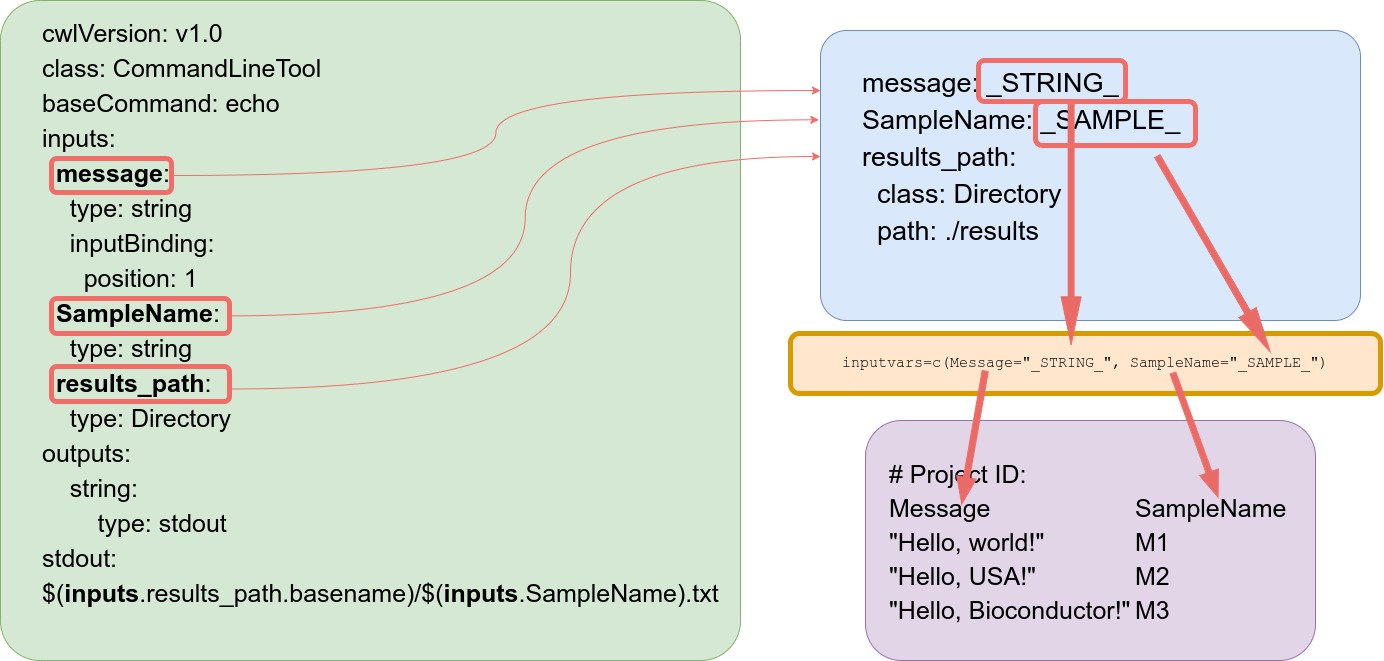
Figure 1: Connectivity between CWL param files and targets files.
Auto-creation of CWL param files from command-line
Users can define the command-line in a pseudo-bash script format. The following used the
the command-line for HISAT2 as example.
command <- "
hisat2 \
-S <F, out: ./results/M1A.sam> \
-x <F: ./data/tair10.fasta> \
-k <int: 1> \
-min-intronlen <int: 30> \
-max-intronlen <int: 3000> \
-threads <int: 4> \
-U <F: ./data/SRR446027_1.fastq.gz>
"
Define prefix and defaults
-
First line is the base command. Each line is an argument with its default value.
-
All following lines specify arguments. Lines starting with a
-or--followed by a non-space delimited letter/word will be interpreted as a prefix, e.g.-Sor--min. Lines without this prefix will be rendered as non-prefix arguments. -
All default settings are placed inside
<...>. Omit for arguments without values such as--verbose. -
First argument is the type of the input.
Ffor “File”, “int” and “string” are unchanged. -
Optional: keyword
outfollowed the type. Separation by,(comma) indicates whether this argument is also a CWL output. -
Use
:to separate keywords and default values. Any non-space separated value after the:will be treated as the default value.
createParamFiles Function
The createParamFiles function accepts as input a command-line provided in above string syntax.
The function returns a cwl with the following components:
BaseCommand: Specifies the program to executeInputs: Defines the input parameters of the processOutputs: Defines the parameters representing the output of the process
The fourth component is the original command-line provided as input.
In interactive mode, the function will verify if everything is correct and
ask the user to proceed. The user can answer “no” and provide more information
at the string input level. Another question is whether to save the generated CWL
results to the corresponding .cwl and .yml files. When running the function
in non-interactive mode, the results will be returned without asking for confirmation
by the user.
cmd <- createParamFiles(command, writeParamFiles = FALSE)
## *****BaseCommand*****
## hisat2
## *****Inputs*****
## S:
## type: File
## preF: -S
## yml: ./results/M1A.sam
## x:
## type: File
## preF: -x
## yml: ./data/tair10.fasta
## k:
## type: int
## preF: -k
## yml: 1
## min-intronlen:
## type: int
## preF: -min-intronlen
## yml: 30
## max-intronlen:
## type: int
## preF: -max-intronlen
## yml: 3000
## threads:
## type: int
## preF: -threads
## yml: 4
## U:
## type: File
## preF: -U
## yml: ./data/SRR446027_1.fastq.gz
## *****Outputs*****
## output1:
## type: File
## value: ./results/M1A.sam
## *****Parsed raw command line*****
## hisat2 -S ./results/M1A.sam -x ./data/tair10.fasta -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz
If the user chooses not to save the param files in the createParamFiles call directly,
then the writeParamFiles function allows to do this in a separate step.
writeParamFiles(cmd, overwrite = TRUE)
## Written content of 'commandLine' to file:
## param/cwl/hisat2/hisat2.cwl
## Written content of 'commandLine' to file:
## param/cwl/hisat2/hisat2.yml
Accessor functions
Print components
Note, the results of createParamFiles are stored in a SYSargs2 container. The individual
components can be accessed as follows.
printParam(cmd, position = "baseCommand") ## Print a baseCommand section
## *****BaseCommand*****
## hisat2
printParam(cmd, position = "outputs")
## *****Outputs*****
## output1:
## type: File
## value: ./results/M1A.sam
printParam(cmd, position = "inputs", index = 1:2) ## Print by index
## *****Inputs*****
## S:
## type: File
## preF: -S
## yml: ./results/M1A.sam
## x:
## type: File
## preF: -x
## yml: ./data/tair10.fasta
printParam(cmd, position = "inputs", index = -1:-2) ## Negative indexing printing to exclude certain indices in a position
## *****Inputs*****
## k:
## type: int
## preF: -k
## yml: 1
## min-intronlen:
## type: int
## preF: -min-intronlen
## yml: 30
## max-intronlen:
## type: int
## preF: -max-intronlen
## yml: 3000
## threads:
## type: int
## preF: -threads
## yml: 4
## U:
## type: File
## preF: -U
## yml: ./data/SRR446027_1.fastq.gz
cmdlist(cmd)
## $defaultid
## $defaultid$hisat2
## [1] "hisat2 -S ./results/M1A.sam -x ./data/tair10.fasta -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz"
Subsetting the command-line
cmd2 <- subsetParam(cmd, position = "inputs", index = 1:2, trim = TRUE)
## *****Inputs*****
## S:
## type: File
## preF: -S
## yml: ./results/M1A.sam
## x:
## type: File
## preF: -x
## yml: ./data/tair10.fasta
## *****Parsed raw command line*****
## hisat2 -S ./results/M1A.sam -x ./data/tair10.fasta
cmdlist(cmd2)
## $defaultid
## $defaultid$hisat2
## [1] "hisat2 -S ./results/M1A.sam -x ./data/tair10.fasta"
cmd2 <- subsetParam(cmd, position = "inputs", index = c("S", "x"), trim = TRUE)
## *****Inputs*****
## S:
## type: File
## preF: -S
## yml: ./results/M1A.sam
## x:
## type: File
## preF: -x
## yml: ./data/tair10.fasta
## *****Parsed raw command line*****
## hisat2 -S ./results/M1A.sam -x ./data/tair10.fasta
cmdlist(cmd2)
## $defaultid
## $defaultid$hisat2
## [1] "hisat2 -S ./results/M1A.sam -x ./data/tair10.fasta"
Replacing existing argument
cmd3 <- replaceParam(cmd, "base", index = 1, replace = list(baseCommand = "bwa"))
## Replacing baseCommand
## *****BaseCommand*****
## bwa
## *****Parsed raw command line*****
## bwa -S ./results/M1A.sam -x ./data/tair10.fasta -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz
cmdlist(cmd3)
## $defaultid
## $defaultid$hisat2
## [1] "bwa -S ./results/M1A.sam -x ./data/tair10.fasta -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz"
new_inputs <- new_inputs <- list(
"new_input1" = list(type = "File", preF="-b", yml ="myfile"),
"new_input2" = "-L <int: 4>"
)
cmd4 <- replaceParam(cmd, "inputs", index = 1:2, replace = new_inputs)
## Replacing inputs
## *****Inputs*****
## new_input1:
## type: File
## preF: -b
## yml: myfile
## new_input2:
## type: int
## preF: -L
## yml: 4
## k:
## type: int
## preF: -k
## yml: 1
## min-intronlen:
## type: int
## preF: -min-intronlen
## yml: 30
## max-intronlen:
## type: int
## preF: -max-intronlen
## yml: 3000
## threads:
## type: int
## preF: -threads
## yml: 4
## U:
## type: File
## preF: -U
## yml: ./data/SRR446027_1.fastq.gz
## *****Parsed raw command line*****
## hisat2 -b myfile -L 4 -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz
cmdlist(cmd4)
## $defaultid
## $defaultid$hisat2
## [1] "hisat2 -b myfile -L 4 -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz"
Adding new arguments
newIn <- new_inputs <- list(
"new_input1" = list(type = "File", preF="-b1", yml ="myfile1"),
"new_input2" = list(type = "File", preF="-b2", yml ="myfile2"),
"new_input3" = "-b3 <F: myfile3>"
)
cmd5 <- appendParam(cmd, "inputs", index = 1:2, append = new_inputs)
## Replacing inputs
## *****Inputs*****
## S:
## type: File
## preF: -S
## yml: ./results/M1A.sam
## x:
## type: File
## preF: -x
## yml: ./data/tair10.fasta
## k:
## type: int
## preF: -k
## yml: 1
## min-intronlen:
## type: int
## preF: -min-intronlen
## yml: 30
## max-intronlen:
## type: int
## preF: -max-intronlen
## yml: 3000
## threads:
## type: int
## preF: -threads
## yml: 4
## U:
## type: File
## preF: -U
## yml: ./data/SRR446027_1.fastq.gz
## new_input1:
## type: File
## preF: -b1
## yml: myfile1
## new_input2:
## type: File
## preF: -b2
## yml: myfile2
## new_input3:
## type: File
## preF: -b3
## yml: myfile3
## *****Parsed raw command line*****
## hisat2 -S ./results/M1A.sam -x ./data/tair10.fasta -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz -b1 myfile1 -b2 myfile2 -b3 myfile3
cmdlist(cmd5)
## $defaultid
## $defaultid$hisat2
## [1] "hisat2 -S ./results/M1A.sam -x ./data/tair10.fasta -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz -b1 myfile1 -b2 myfile2 -b3 myfile3"
cmd6 <- appendParam(cmd, "inputs", index = 1:2, after=0, append = new_inputs)
## Replacing inputs
## *****Inputs*****
## new_input1:
## type: File
## preF: -b1
## yml: myfile1
## new_input2:
## type: File
## preF: -b2
## yml: myfile2
## new_input3:
## type: File
## preF: -b3
## yml: myfile3
## S:
## type: File
## preF: -S
## yml: ./results/M1A.sam
## x:
## type: File
## preF: -x
## yml: ./data/tair10.fasta
## k:
## type: int
## preF: -k
## yml: 1
## min-intronlen:
## type: int
## preF: -min-intronlen
## yml: 30
## max-intronlen:
## type: int
## preF: -max-intronlen
## yml: 3000
## threads:
## type: int
## preF: -threads
## yml: 4
## U:
## type: File
## preF: -U
## yml: ./data/SRR446027_1.fastq.gz
## *****Parsed raw command line*****
## hisat2 -b1 myfile1 -b2 myfile2 -b3 myfile3 -S ./results/M1A.sam -x ./data/tair10.fasta -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz
cmdlist(cmd6)
## $defaultid
## $defaultid$hisat2
## [1] "hisat2 -b1 myfile1 -b2 myfile2 -b3 myfile3 -S ./results/M1A.sam -x ./data/tair10.fasta -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz"
Editing output param
new_outs <- list(
"sam_out" = "<F: $(inputs.results_path)/test.sam>"
)
cmd7 <- replaceParam(cmd, "outputs", index = 1, replace = new_outs)
## Replacing outputs
## *****Outputs*****
## sam_out:
## type: File
## value: $(inputs.results_path)/test.sam
## *****Parsed raw command line*****
## hisat2 -S ./results/M1A.sam -x ./data/tair10.fasta -k 1 -min-intronlen 30 -max-intronlen 3000 -threads 4 -U ./data/SRR446027_1.fastq.gz
output(cmd7)
## $defaultid
## $defaultid$hisat2
## [1] "./results/test.sam"
Version information
sessionInfo()
## R version 4.4.0 (2024-04-24)
## Platform: x86_64-pc-linux-gnu
## Running under: Debian GNU/Linux 11 (bullseye)
##
## Matrix products: default
## BLAS: /usr/lib/x86_64-linux-gnu/blas/libblas.so.3.9.0
## LAPACK: /usr/lib/x86_64-linux-gnu/lapack/liblapack.so.3.9.0
##
## locale:
## [1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
## [3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
## [5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
## [7] LC_PAPER=en_US.UTF-8 LC_NAME=C
## [9] LC_ADDRESS=C LC_TELEPHONE=C
## [11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
##
## time zone: America/Los_Angeles
## tzcode source: system (glibc)
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] systemPipeR_2.10.0 ShortRead_1.62.0
## [3] GenomicAlignments_1.40.0 SummarizedExperiment_1.34.0
## [5] Biobase_2.64.0 MatrixGenerics_1.16.0
## [7] matrixStats_1.3.0 BiocParallel_1.38.0
## [9] Rsamtools_2.20.0 Biostrings_2.72.0
## [11] XVector_0.44.0 GenomicRanges_1.56.0
## [13] GenomeInfoDb_1.40.0 IRanges_2.38.0
## [15] S4Vectors_0.42.0 BiocGenerics_0.50.0
##
## loaded via a namespace (and not attached):
## [1] gtable_0.3.5 xfun_0.43 bslib_0.7.0
## [4] hwriter_1.3.2.1 ggplot2_3.5.1 htmlwidgets_1.6.4
## [7] latticeExtra_0.6-30 lattice_0.22-6 generics_0.1.3
## [10] vctrs_0.6.5 tools_4.4.0 bitops_1.0-7
## [13] parallel_4.4.0 fansi_1.0.6 tibble_3.2.1
## [16] pkgconfig_2.0.3 Matrix_1.7-0 RColorBrewer_1.1-3
## [19] lifecycle_1.0.4 GenomeInfoDbData_1.2.12 stringr_1.5.1
## [22] compiler_4.4.0 deldir_2.0-4 munsell_0.5.1
## [25] codetools_0.2-20 htmltools_0.5.8.1 sass_0.4.9
## [28] yaml_2.3.8 pillar_1.9.0 crayon_1.5.2
## [31] jquerylib_0.1.4 DelayedArray_0.30.0 cachem_1.0.8
## [34] abind_1.4-5 tidyselect_1.2.1 digest_0.6.35
## [37] stringi_1.8.3 dplyr_1.1.4 bookdown_0.39
## [40] fastmap_1.1.1 grid_4.4.0 colorspace_2.1-0
## [43] cli_3.6.2 SparseArray_1.4.0 magrittr_2.0.3
## [46] S4Arrays_1.4.0 utf8_1.2.4 UCSC.utils_1.0.0
## [49] scales_1.3.0 rmarkdown_2.26 pwalign_1.0.0
## [52] httr_1.4.7 jpeg_0.1-10 interp_1.1-6
## [55] blogdown_1.19 png_0.1-8 evaluate_0.23
## [58] knitr_1.46 rlang_1.1.3 Rcpp_1.0.12
## [61] glue_1.7.0 jsonlite_1.8.8 R6_2.5.1
## [64] zlibbioc_1.50.0
Funding
This project is funded by NSF award ABI-1661152.
References
Amstutz, Peter, Michael R Crusoe, Nebojša Tijanić, Brad Chapman, John Chilton, Michael Heuer, Andrey Kartashov, et al. 2016. “Common Workflow Language, V1.0,” July. https://doi.org/10.6084/m9.figshare.3115156.v2.