Generate list of CEL names defining treatment vs. control comparisons
The sampleList function extracts the sample comparisons (contrasts) from the
CMAP annotation table and stores them as a list.
cmap <- read.delim("./data/cmap_instances_02.txt", check.names=FALSE)
# comp_list <- sampleList(cmap, myby="CMP")
comp_list <- sampleList(cmap, myby="CMP_CELL")
Load normalized expression data
The following loads the MAS5 normalized expression data into a single data.frame.
To accelerate the import, the data is read from rds files.
chiptype_dir <- unique(readRDS("./data/chiptype.rds"))
df1 <- readRDS(paste0("data/", chiptype_dir[1], "/", "all_mas5exprs.rds"))
df2 <- readRDS(paste0("data/", chiptype_dir[2], "/", "all_mas5exprs.rds"))
df3 <- readRDS(paste0("data/", chiptype_dir[3], "/", "all_mas5exprs.rds"))
affyid <- rownames(df1)[rownames(df1) %in% rownames(df2)]; affyid <- affyid[affyid %in% rownames(df3)]
mas5df <- cbind(df1[affyid,], df2[affyid,], df3[affyid,])
The next step generates gene level expression values. If genes are represented by several
probe sets then their mean intensities are used. This is necessary because
the U133 chip contains many genes with duplicated probe sets. Probe sets not matching
any gene are removed.
myAnnot <- readRDS("./results/myAnnot.rds")
myAnnot <- myAnnot[as.character(myAnnot[,"ENTREZID"]) != "NA",]
mas5df <- mas5df[rownames(myAnnot),]
idlist <- tapply(row.names(myAnnot), as.character(myAnnot$ENTREZID), c)
mas5df <- t(sapply(names(idlist), function(x) colMeans(mas5df[idlist[[x]], ])))
DEG analysis with limma
The analysis of differentially expressed genes (DEGs) is performed with the limma package.
Genes meeting the chosen cutoff criteria are reported as DEGs (below set to FDR of 10% and
a minimum fold change of 2). The DEG matrix is written to a file named
degMA.xls.
degList <- runLimma(df=log2(mas5df), comp_list, fdr=0.10, foldchange=1, verbose=TRUE, affyid=NULL)
write.table(degList$DEG, file="./results/degMA.xls", quote=FALSE, sep="\t", col.names = NA)
saveRDS(degList$DEG, "./results/degMA.rds") # saves binary matrix
saveRDS(degList, "./results/degList.rds") # saves entire degList
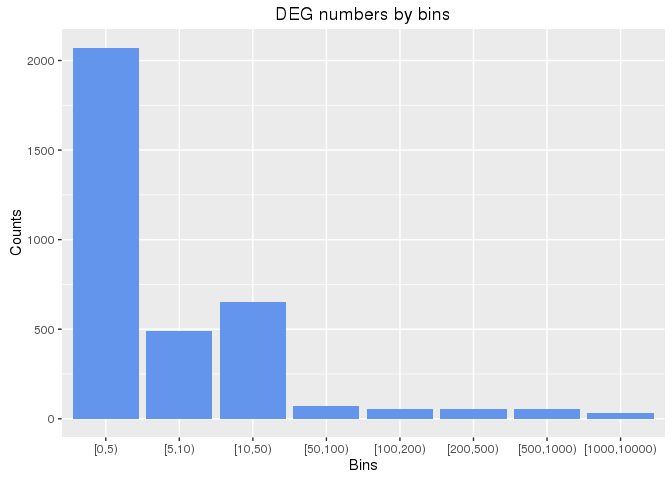
Number of DEGs across drug treatments
The following plots the number of drug treatments (y-axis) for increasing bin sizes (x-axis)
of DEGs.
degMAgene <- readRDS("./results/degMA.rds")
y <- as.numeric(colSums(degMAgene))
interval <- table(cut(y, right=FALSE, dig.lab=5, breaks=c(0, 5, 10, 50, 100, 200, 500, 1000, 10000)))
df <- data.frame(interval); colnames(df) <- c("Bins", "Counts")
ggplot(df, aes(Bins, Counts)) +
geom_bar(position="dodge", stat="identity", fill="cornflowerblue") +
ggtitle("DEG numbers by bins")
Identify DEG overlaps with Peters et al. (2015)
Peters et al. (2015) reported 1,497 age-related gene expression
signatures. The intersectStats function computes their intersects with each
of the 3,318 drug-responsive DEG sets from CMAP. The result includes the
Jaccard index as a simple similarity metric for gene sets as well as the raw
and adjusted p-values based on the hypergeometric distribution expressing how
likely it is to obtain the observed intersect sizes just by chance. The
results for the 20 top scoring drugs are given below and the full data set is
written to a file named
degOL_PMID26490707.xls.
PMID26490707 <- read.delim("./data/PMID26490707_S1.xls", comment="#")
myAnnot <- readRDS("./results/myAnnot.rds")
geneid <- as.character(PMID26490707$"NEW.Entrez.ID")
degMAgene <- readRDS("./results/degMA.rds") # Faster than read.delim()
degMAsub <- degMAgene[rownames(degMAgene) %in% geneid,]
degOL_PMID26490707 <- intersectStats(degMAgene, degMAsub)
write.table(degOL_PMID26490707, file="./results/degOL_PMID26490707.xls", quote=FALSE, sep="\t", col.names = NA)
sum(degOL_PMID26490707[,1] > 0) # Drugs with any overlap
## [1] 1137
degOL_PMID26490707[1:20,]
## Jaccard_Index longevity_DEG cmap_DEG Intersect Pval
## alprostadil_HL60 0.10272537 1192 912 196 1.706881e-29
## dihydroergotamine_HL60 0.06187625 1192 404 93 3.698500e-16
## pergolide_HL60 0.07815080 1192 767 142 2.631561e-15
## (-)-isoprenaline_HL60 0.05113636 1192 288 72 1.087773e-14
## Prestwick-983_HL60 0.05948447 1192 411 90 2.785586e-14
## nocodazole_HL60 0.06384324 1192 491 101 4.863951e-14
## anisomycin_HL60 0.09790419 1192 2475 327 2.312553e-11
## tetryzoline_HL60 0.03366488 1192 159 44 4.993036e-11
## mebendazole_HL60 0.06041924 1192 528 98 6.663275e-11
## methylergometrine_HL60 0.04436620 1192 291 63 3.825164e-10
## lycorine_HL60 0.09093994 1192 2023 268 2.411922e-09
## podophyllotoxin_HL60 0.04175513 1192 280 59 4.108204e-09
## colchicine_HL60 0.03181818 1192 170 42 5.960591e-09
## co-dergocrine mesilate_HL60 0.05239617 1192 455 82 1.062344e-08
## puromycin_HL60 0.05060858 1192 448 79 5.282895e-08
## tretinoin_HL60 0.03738318 1192 251 52 6.182212e-08
## 15-delta prostaglandin J2_HL60 0.04560698 1192 367 68 6.583181e-08
## geldanamycin_HL60 0.03068862 1192 185 41 2.331048e-07
## bromocriptine_HL60 0.02739726 1192 158 36 6.194257e-07
## apigenin_HL60 0.06292967 1192 818 119 1.601978e-06
## adj_Pval
## alprostadil_HL60 5.936533e-26
## dihydroergotamine_HL60 1.286338e-12
## pergolide_HL60 9.152569e-12
## (-)-isoprenaline_HL60 3.783276e-11
## Prestwick-983_HL60 9.688270e-11
## nocodazole_HL60 1.691682e-10
## anisomycin_HL60 8.043059e-08
## tetryzoline_HL60 1.736578e-07
## mebendazole_HL60 2.317487e-07
## methylergometrine_HL60 1.330392e-06
## lycorine_HL60 8.388664e-06
## podophyllotoxin_HL60 1.428833e-05
## colchicine_HL60 2.073093e-05
## co-dergocrine mesilate_HL60 3.694832e-05
## puromycin_HL60 1.837391e-04
## tretinoin_HL60 2.150173e-04
## 15-delta prostaglandin J2_HL60 2.289630e-04
## geldanamycin_HL60 8.107384e-04
## bromocriptine_HL60 2.154363e-03
## apigenin_HL60 5.571679e-03
Identify DEG overlaps with Sood et al. (2015)
Sood et al. (2015) reported 150 age-related gene expression signatures.
The intersectStats function computes their intersects with each of the 3,318
drug-responsive DEG sets from CMAP. The result includes the Jaccard index as a simple
similarity metric for gene sets as well as the raw and adjusted p-values based on the
hypergeometric distribution expressing how likely it is to observe the observed intersect
sizes just by chance. The results for the 20 top scoring drugs are given below and the full
data set is written to a file named degOL_PMID26343147.xls.
PMID26343147 <- read.delim("./data/PMID26343147_S1T1.xls", check.names=FALSE, comment="#")
myAnnot <- readRDS("./results/myAnnot.rds")
geneid <- as.character(myAnnot[rownames(myAnnot) %in% as.character(PMID26343147[,1]), "ENTREZID"])
geneid <- geneid[geneid!="NA"]
degMA <- readRDS("./results/degMA.rds") # Faster then read.delim()
degMA <- degMA[ , !is.na(colSums(degMA))] # Remove columns where DEG analysis was not possible
degMAsub <- degMA[geneid,]
degOL_PMID26343147 <- intersectStats(degMAgene, degMAsub)
write.table(degOL_PMID26343147, file="./results/degOL_PMID26343147.xls", quote=FALSE, sep="\t", col.names = NA)
sum(degOL_PMID26343147[,1] > 0) # Drugs with any overlap
## [1] 223
degOL_PMID26343147[1:20,] # Top 20 scoring drugs
## Jaccard_Index longevity_DEG cmap_DEG Intersect Pval adj_Pval
## colecalciferol_MCF7 0.017857143 55 2 1 0.008821835 1
## esculin_HL60 0.017857143 55 2 1 0.008821835 1
## flecainide_MCF7 0.017857143 55 2 1 0.008821835 1
## ribostamycin_MCF7 0.017857143 55 2 1 0.008821835 1
## withaferin A_MCF7 0.016000000 55 199 4 0.011470972 1
## dexamethasone_HL60 0.022222222 55 37 2 0.011573654 1
## lobeline_HL60 0.007987220 55 1838 15 0.011600566 1
## idoxuridine_MCF7 0.017543860 55 3 1 0.013204054 1
## metronidazole_MCF7 0.017543860 55 3 1 0.013204054 1
## sulpiride_MCF7 0.017543860 55 3 1 0.013204054 1
## ampyrone_HL60 0.008880995 55 1081 10 0.018769135 1
## chlortetracycline_HL60 0.008080808 55 1442 12 0.021597597 1
## estropipate_MCF7 0.016949153 55 5 1 0.021911500 1
## flunixin_PC3 0.016949153 55 5 1 0.021911500 1
## sulfafurazole_HL60 0.018867925 55 53 2 0.022894704 1
## (-)-MK-801_MCF7 0.016666667 55 6 1 0.026236889 1
## carbarsone_PC3 0.016666667 55 6 1 0.026236889 1
## famprofazone_HL60 0.016666667 55 6 1 0.026236889 1
## guanethidine_HL60 0.016666667 55 6 1 0.026236889 1
## cefalotin_MCF7 0.016393443 55 7 1 0.030543497 1
Drugs affecting known longevity genes
The following identifies CMAP drugs affecting the expression of the IGF1 or IGF1R genes.
The final result is written to a file named deg_IGF1.xls.
genesymbols <- c("IGF1", "IGF1R")
geneids <- unique(as.character(myAnnot[myAnnot$SYMBOL %in% genesymbols,"ENTREZID"]))
names(geneids) <- unique(as.character(myAnnot[myAnnot$SYMBOL %in% genesymbols,"SYMBOL"]))
degList <- readRDS("./results/degList.rds")
df <- data.frame(row.names=colnames(degList$DEG), check.names=FALSE)
index <- which(colSums(degList$DEG[geneids,])>= 1)
for(i in seq_along(geneids)) {
tmp <- data.frame(DEG=degList$DEG[geneids[i],index], logFC=degList$logFC[geneids[i],index], FDR=degList$FDR[geneids[i],index])
colnames(tmp) <- paste0(names(geneids)[i], "_", colnames(tmp))
df <- cbind(df, tmp[rownames(df),] )
}
df <- df[names(index),]
write.table(df, file="./results/deg_IGF1.xls", quote=FALSE, sep="\t", col.names = NA)
Now the final data.frame can be sorted by increasing mean FDR values.
igfDF <- read.delim("./results/deg_IGF1.xls", row.names=1)
igfDF[order(rowMeans(igfDF[,c(3,6)])),][1:20,]
## IGF1R_DEG IGF1R_logFC IGF1R_FDR IGF1_DEG IGF1_logFC IGF1_FDR
## camptothecin_MCF7 1 -1.48180690 4.867357e-07 0 0.94295576 0.07821138
## 0175029-0000_MCF7 0 -0.62026588 1.193479e-01 1 -1.01210916 0.04295254
## cicloheximide_HL60 1 -1.29607754 2.054384e-02 0 1.67183429 0.16908728
## emetine_HL60 1 -1.23843896 3.085463e-03 0 1.03812083 0.26576009
## digoxigenin_PC3 0 0.54850849 2.488776e-01 1 -1.27142976 0.04219010
## anisomycin_HL60 1 -2.85399433 1.481347e-03 0 0.66701644 0.44025263
## vorinostat_MCF7 1 -1.50640830 3.143781e-17 0 0.37179771 0.45437384
## irinotecan_MCF7 1 -2.26588384 9.895790e-02 0 -1.05446828 0.41746319
## tyrphostin AG-825_MCF7 1 -1.03570748 3.440048e-02 0 1.57208567 0.51400841
## trichostatin A_MCF7 1 -1.27541131 6.943847e-127 0 0.07335895 0.55046789
## baclofen_HL60 1 1.14379132 8.226627e-02 0 -1.00641253 0.53322529
## 8-azaguanine_PC3 1 -1.04668136 7.466879e-02 0 -0.97679589 0.65221001
## verteporfin_MCF7 1 -1.11136304 1.226251e-03 0 0.64194152 0.78463681
## piperlongumine_MCF7 1 -1.03214332 1.088011e-02 0 0.53820854 0.78914488
## verteporfin_HL60 1 -1.17232840 4.923492e-02 0 1.09438523 0.75358370
## cephaeline_HL60 1 -1.13448661 1.479635e-02 0 -0.19882135 0.86619935
## lycorine_HL60 1 -1.93807396 1.390079e-03 0 -0.23062131 0.88748758
## doxorubicin_MCF7 1 -1.08389932 4.035039e-04 0 0.12613722 0.92452703
## anisomycin_MCF7 0 0.06265744 8.938603e-01 1 -1.55915713 0.04355888
## 15-delta prostaglandin J2_MCF7 0 0.01396391 9.774076e-01 1 1.03626957 0.09233156
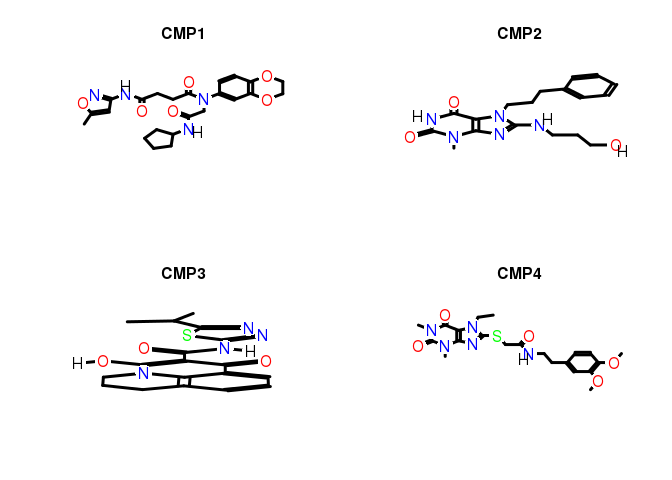
Plot structures of compounds
library(ChemmineR)
mypath <- system.file("extdata", "longevitydrugs.sdf", package="longevityTools")
mypath <- "../inst/extdata/longevitydrugs.sdf"
sdfset <- read.SDFset(mypath)
data(sdfsample)
sdfsample
## An instance of "SDFset" with 100 molecules
plot(sdfsample[1:4], print=FALSE)