Authors: Kevin Horan, Yiqun Cao, Tyler Backman, Thomas Girke
Last update: 06 March, 2016
Alternative formats of this tutorial:
.Rmd HTML,
.Rmd Source,
.R Source,
PDF Slides
Introduction
ChemmineR is a cheminformatics package for analyzing
drug-like small molecule data in R. Its latest version contains
functions for efficient processing of large numbers of small molecules,
physicochemical/structural property predictions, structural similarity
searching, classification and clustering of compound libraries with a
wide spectrum of algorithms.
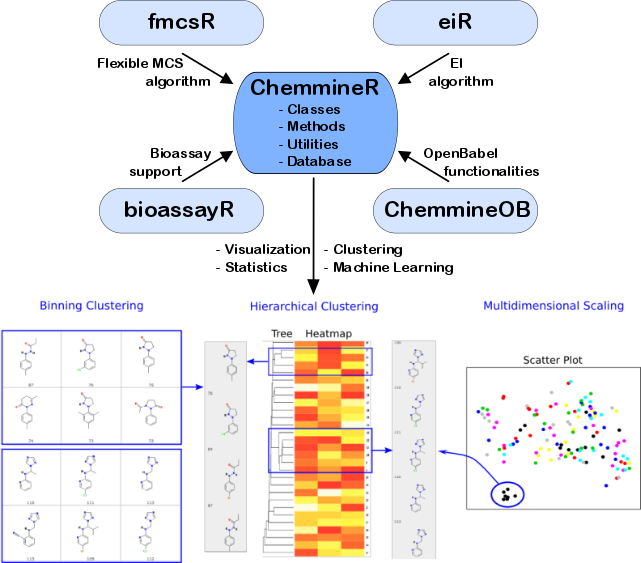
In addition, ChemmineR offers visualization functions
for compound clustering results and chemical structures. The integration
of chemoinformatic tools with the R programming environment has many
advantages, such as easy access to a wide spectrum of statistical
methods, machine learning algorithms and graphic utilities. The first
version of this package was published in Cao et al. (2008). Since then many additional
utilities and add-on packages have been added to the environment (Figure 2) and
many more are under development for future releases (T. W. H. Backman et al., 2011; Y. Wang et al., 2013).
Recently Added Features
-
Improved SMILES support via new SMIset object class
and SMILES import/export functions
-
Integration of a subset of OpenBabel functionalities via new
ChemmineOB add-on package (Y. Cao et al., 2008)
-
Streaming functionality for processing millions of molecules on a
laptop
-
Mismatch tolerant maximum common substructure (MCS) search algorithm
-
Fast and memory efficient fingerprint search support using atom pair
or PubChem fingerprints